Phylogenetics
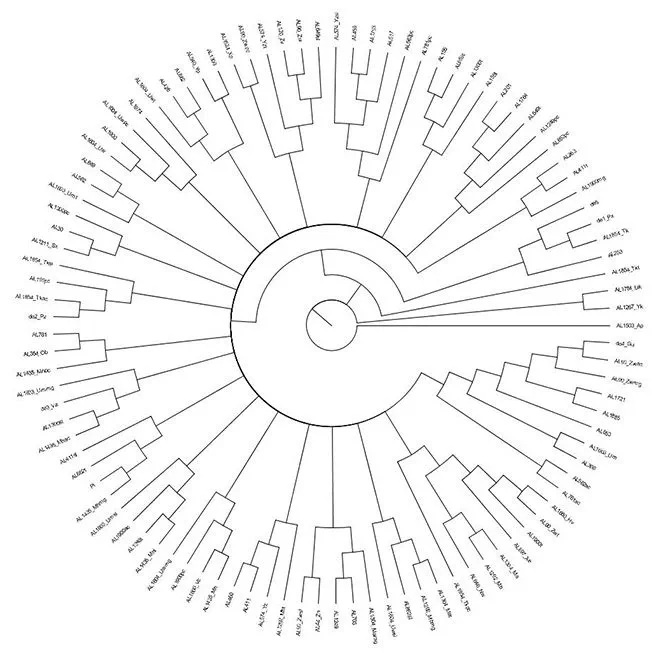
As a follow-up to my digital critical edition of Pietro d’Abano’s commentary, I am creating an edition of the Latin translation of the text he commented on: pseudo-Aristotle’s Problemata. Since there are 50 surviving medieval manuscripts that preserve this text, traditional methods for establishing a stemma (a diagram reconstructing the manuscripts’ familial relationships) are not feasible. Instead, I have quantified the manuscripts’ degree of relatedness by using tools from the field of phylogenetics, which aims to reconstruct the evolutionary history of a given species.
Undertaking phylogenetic-based text criticism entails entering the textual variants found in representative passages from all 50 manuscripts into a TEI-compliant XML document. I then use teiphy, a Python package that converts TEI XML into the .nexus file format used in phylogenetics. That file is then readable by PAUP*, a program that conducts phylogenetic analyses according to a variety of criteria, including parsimony and likelihood. The resulting analyses can be mapped in numerous ways, such as is shown in the image above.